Tanjeem Azwad Zaman
|
Ph.D. Student
Carnegie Mellon University, SCS
About me
I'm Tanjeem, a Ph.D. student (Fall'25) in the Joint CMU-Pitt PhD Program in Computational Biology, under Carnegie Mellon University's School of Computer Science. I completed my undergraduate from the Department of Computer Science & Engineering (CSE) at Bangladesh University of Engineering and Technology (BUET). My research interests lies in Bioinformatics, Algorithms and Machine Learning.
Outside of my academic pursuits, I am an avid fan of problem-solving -- which would explain my fascination with Olympiads during middle and high school. In my free time, I enjoy playing soccer, watching anime, or covering OSTs on the violin.
Work Experience
-
Lecturer
Department of Computer Science and Engineering,
BRAC University
January 2025 — July 2025[CSE331] Automata and Computability (Theory)
[CSE423] Computer Graphics (Theory + Lab)
-
Adjunct Lecturer
Department of Computer Science and Engineering,
Bangladesh University of Engineering and Technology
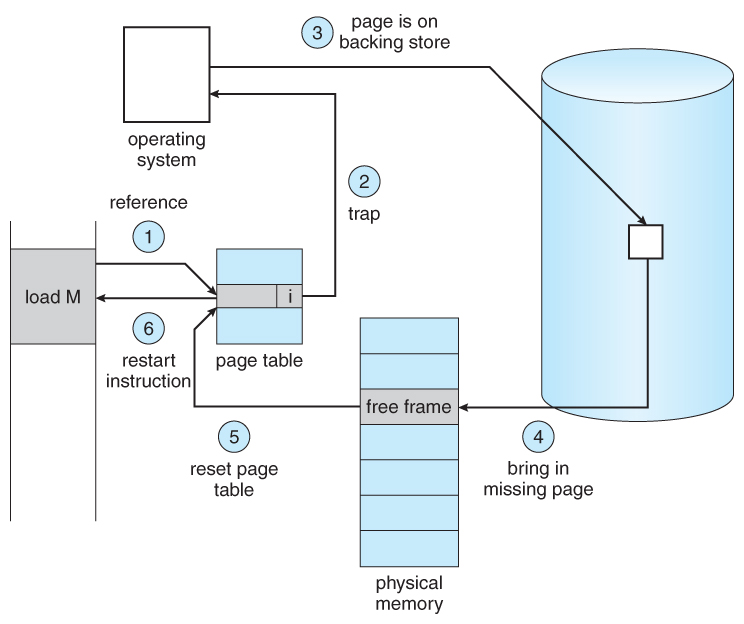
August 2024 — December 2024[CSE211] Theory of Computation (Theory)
[CSE326] Information Systems Development and Management (Lab)
[CSE322] Computer Networks (Lab)
[CSE210] Computer Architecture (Lab)
[CSE200] Technical Writing and Presentation (Lab)
-
Machine Learning Intern
North-West Power Generation Company Limited, Bangladesh
April 2023 — June 2023Part of an internship program organized by the Department of CSE, BUET. Collected data from several sensors installed around the NWPGCL plant and used ML models to predict the operational state of machinery.
-
Software Development Intern
May 2023 — June 2023I was part of a team in charge of developing an app with real-time data analysis and visualization for a foreign client. Built a full-stack module from scratch: Data-Analysis (pandas), Backend (FastAPI), and Frontend development (JavaScript).
Publications and Research Experience
-
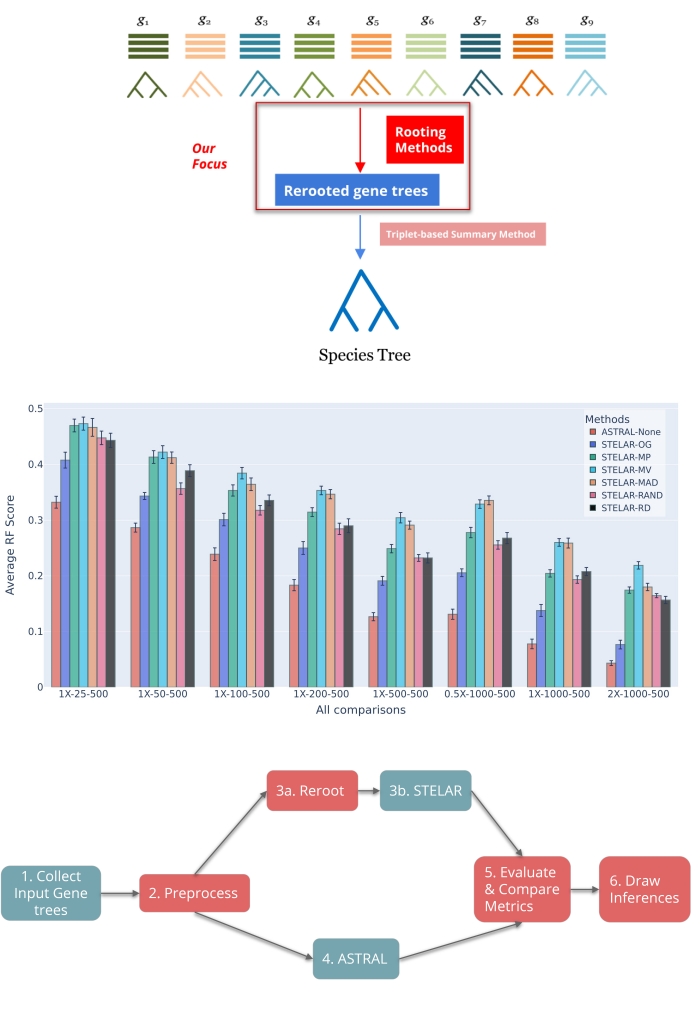
Robustness to Gene Tree Rooting of Triplet-based Species Tree Estimation Methods (August 2023 - March 2025)
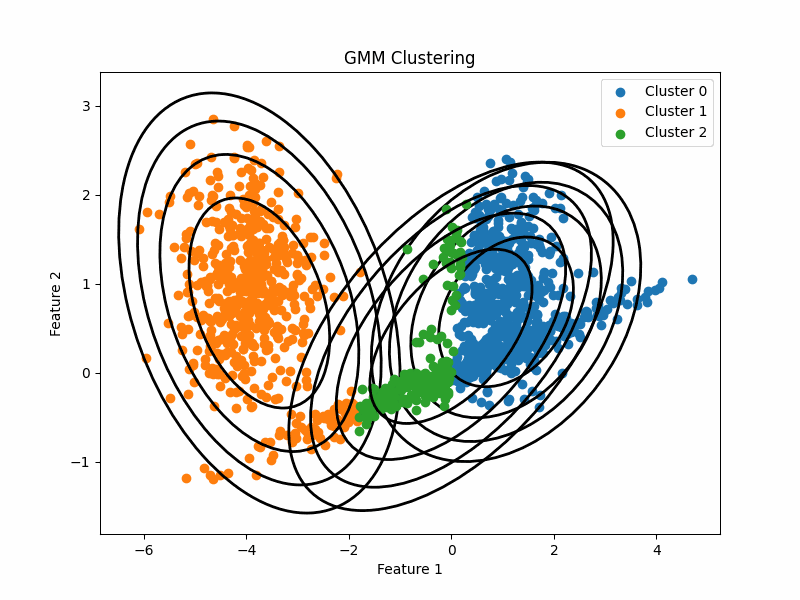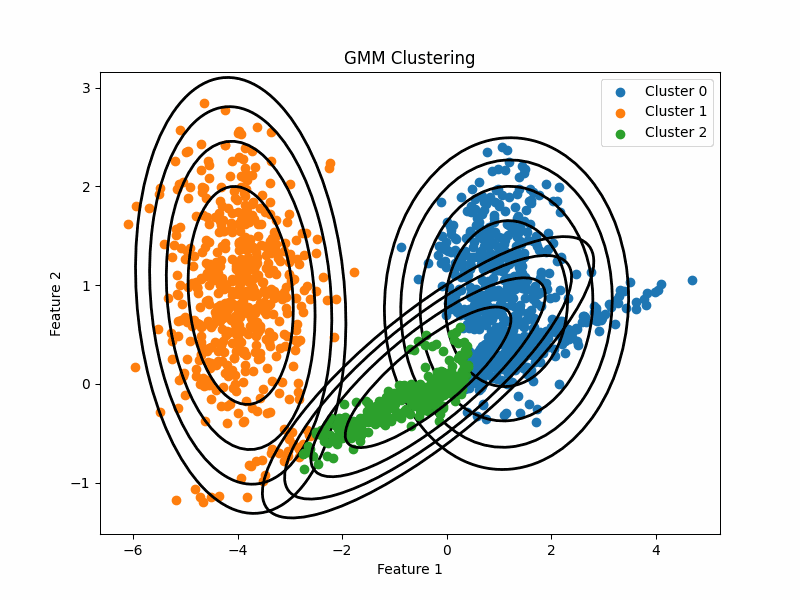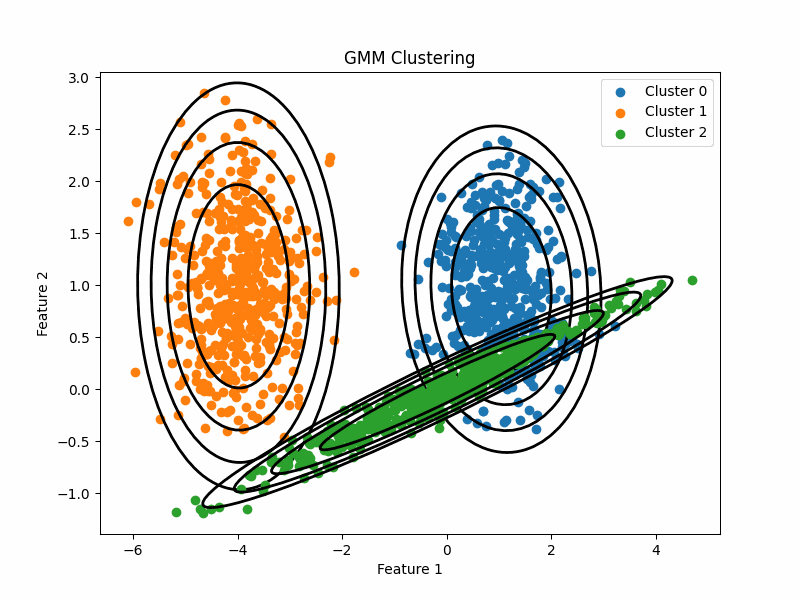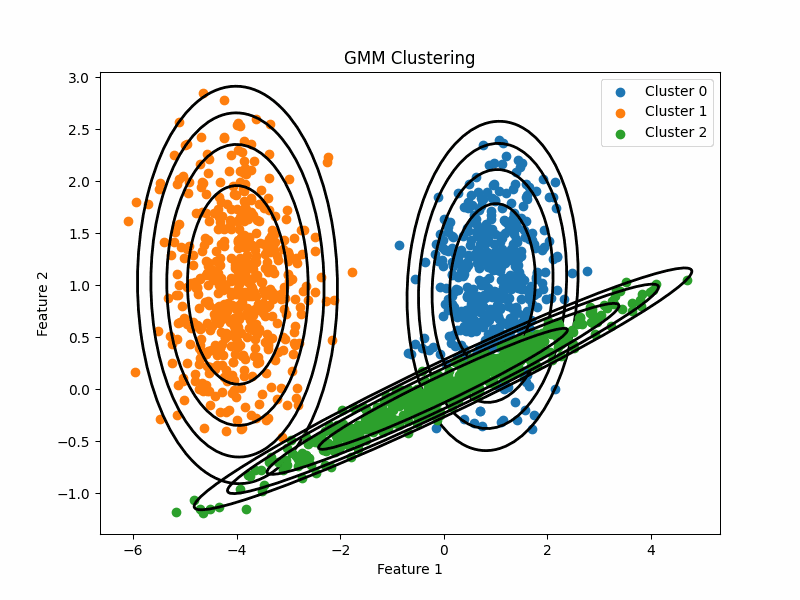
Computational Biology, Phylogenetics Published at RECOMB-CG 2025 Supervised by Dr. Md Shamsuzzoha BayzidWe conducted rigorous empirical testing with different phylogenetic rooting techniques on input gene trees to evaluate their effect on the species trees estimated by triplet-based estimation methods like STELAR. We evaluated key parameters such as RF Distance, Triplet and Quartet Scores for simulated datasets with varying levels of Incomplete Lineage Sorting (ILS), gene counts, and base pairs, and considered the correctness of inter-clade relationships for biological datasets. Our analysis identified and explained conditions where STELAR with algorithmic rootings yielded better performance than with Outgroup rooting, even outperforming the state-of-the-art species tree estimation method ASTRAL. Finally, we suggested an avenue for the underrated triplet-based methods to excel even when the true root is unknown. DOI: https://doi.org/10.1007/978-3-031-94928-9_8
-
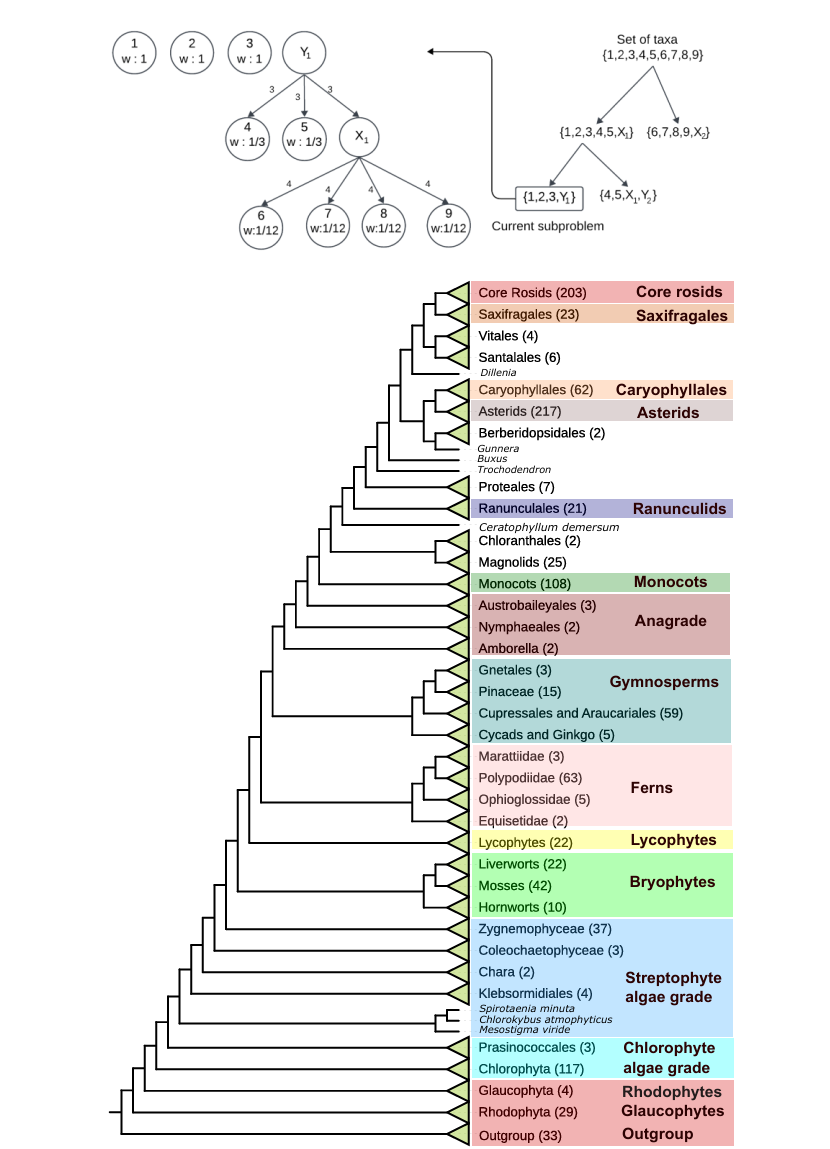
wQFM-TREE: Highly Accurate and Scalable Quartet-Based Species Tree Inference from Gene Trees (August 2023 - July 2024)
Published at Bioinformatics Advances Computational Biology, Phylogenetics, Algorithms Supervised by Dr. Md Shamsuzzoha BayzidWe addressed the scalability issues of the quartet-based species tree estimation method wQFM. We introduced wQFM-TREE, which uses graph theory, combinatorics, and data structures efficiently to avoid explicitly enumerating all possible quartets, as done in wQFM. Our method was benchmarked against other state-of-the-art (SOTA) methods on simulated datasets (up to 2000 taxa) and empirical datasets (the One Thousand Plant Transcriptomes Initiative dataset). The results showed that wQFM-TREE consistently outperformed or matched the accuracy of SOTA methods, ASTRAL and TREE-QMC, especially in cases with higher taxa, showcasing its competitiveness in large-scale phylogenetic analyses. DOI: https://doi.org/10.1093/bioadv/vbaf053
-
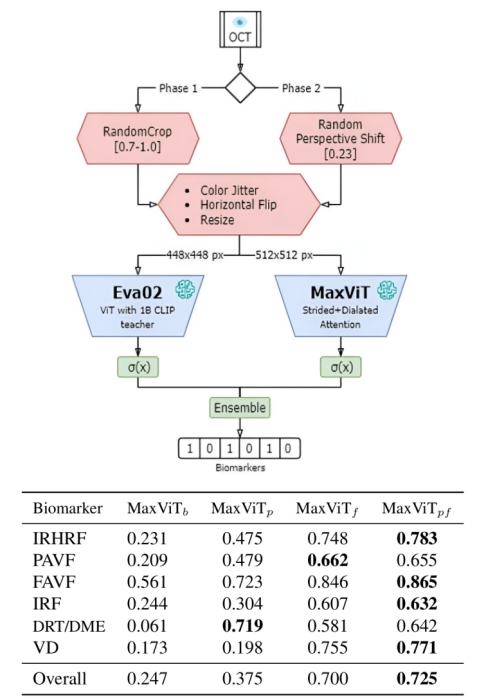
Ophthalmic Biomarker Detection using Ensembled Vision Transformers (October 2023 - March 2025)
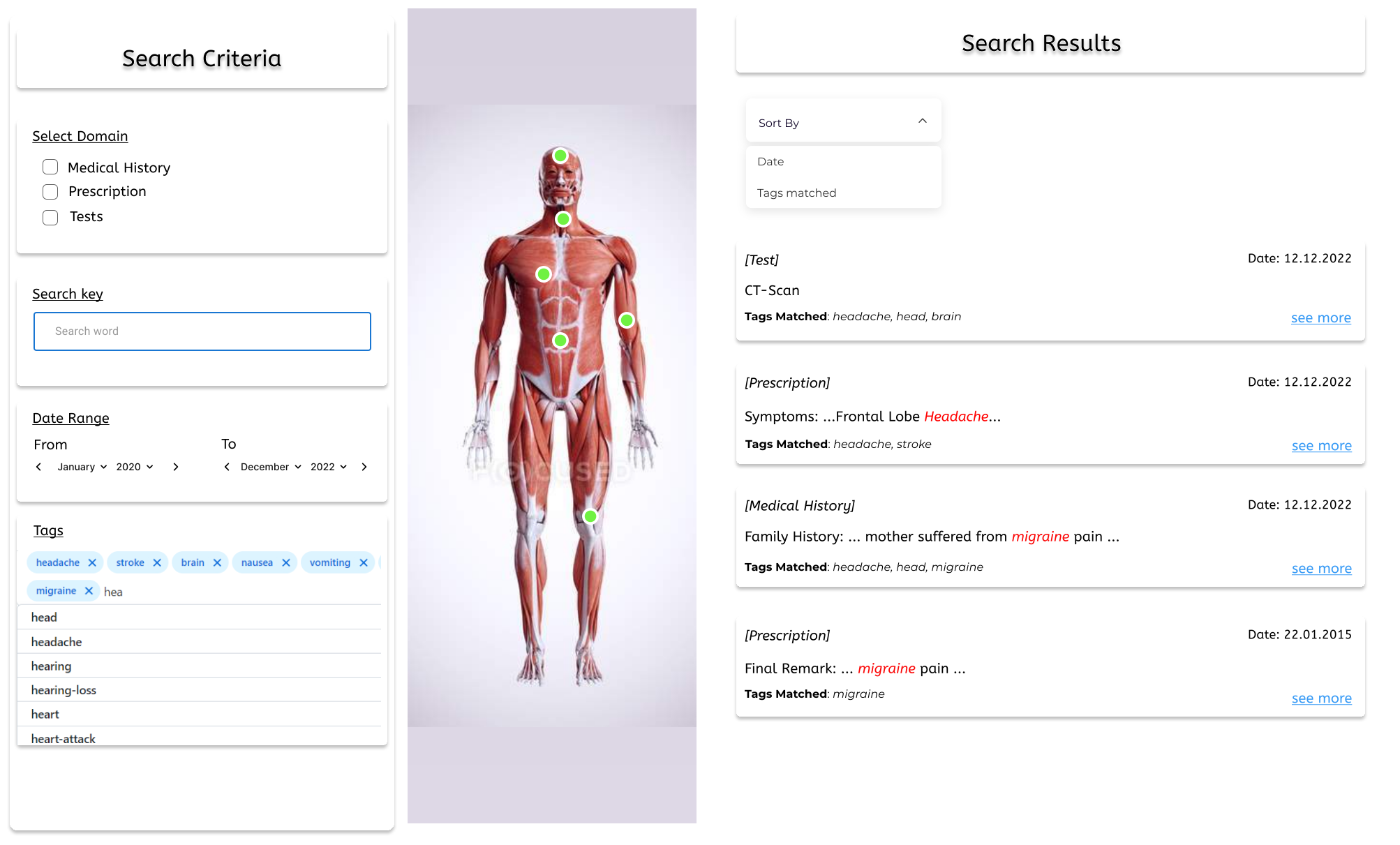
Medical Imaging, Machine Learning Supervised by Dr. M. Sohel RahmanWe used extensive data augmentations and meticulous ensembling of vision transformers to detect ophthalmic biomarkers from OCT scans, resulting in placing 1st at the IEEE SPS VIP Cup 2023. We consulted an ophthalmologist to understand both local and global features, and ensembled MaxViT (which utilizes strided attention to focus on local features) and EVA-02 (which excels at detecting global features) to form the winning idea. During training, we employed various data augmentation strategies including random greyscale transformation, color jitter, random resized cropping, horizontal flipping, and random perspective shifts to enhance model robustness. Post-competition, we utilized unlabelled data by adopting pseudolabeling and knowledge distillation techniques to train a single MaxViT model that matched our winning solution while significantly reducing inference time and computational cost. Preprint available at: https://doi.org/10.48550/arXiv.2310.14005
-
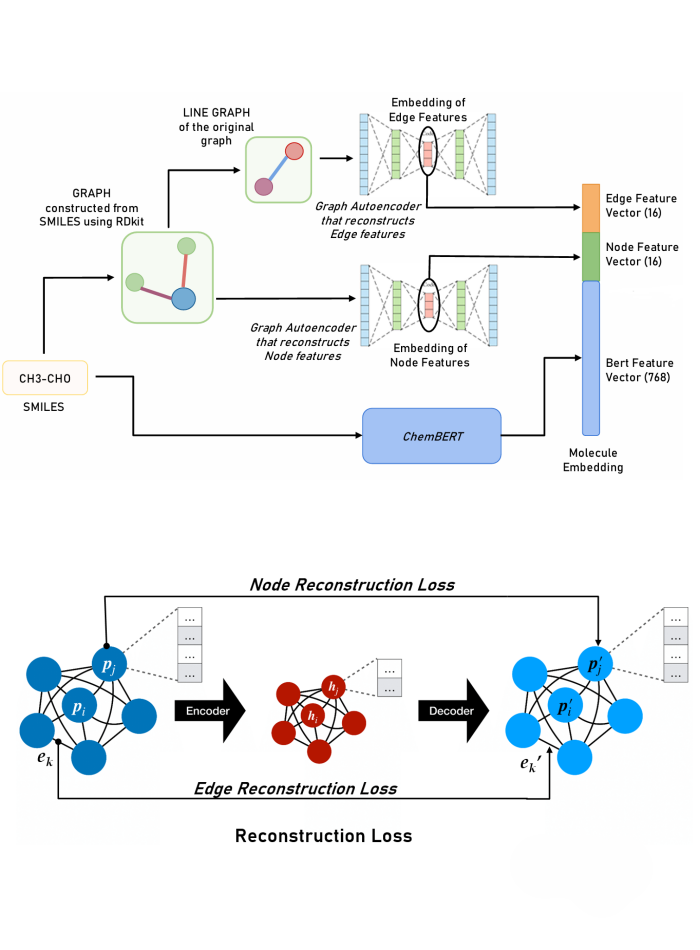
Dual Graph Autoencoders for Enhanced Graph Representations (November 2023 - May 2025)
Cheminformatics, Machine Learning, Graph Neural Networks Supervised by Dr. Mohammad Saifur RahmanWe developed a Dual-Graph Autoencoder framework that effectively utilizes both node features and the traditionally overlooked edge features in graph-structured data. Graphs were made using node and edge features from RDKit, and final features were composed of latent layers of separate autoencoders trained on the graphs and their line-graphs, along with embeddings extracted by ChemBERT from SMILES. The framework was benchmarked on the Tox-21 dataset (a multi-label binary classification task) and the HIV dataset from MoleculeNet, where our approach outperformed other GNN-based methods that used edge features, such as Graph Transformers. We also conducted ablation studies using different feature combinations, providing insights into the efficiency of structural versus semantic features in each classification task. Preprint available at: https://doi.org/10.1101/2024.11.22.624875
Education
-
Ph.D. in Computational Biology
School of Computer Science, Carnegie Mellon University
August 2025 - Present -
B.Sc. in Computer Science and Engineering
Bangladesh University of Engineering and Technology
April 2019 - July 2024CGPA: 4.00/4.00
- Ranked 1st in a class of 113 students
Notable Courses:
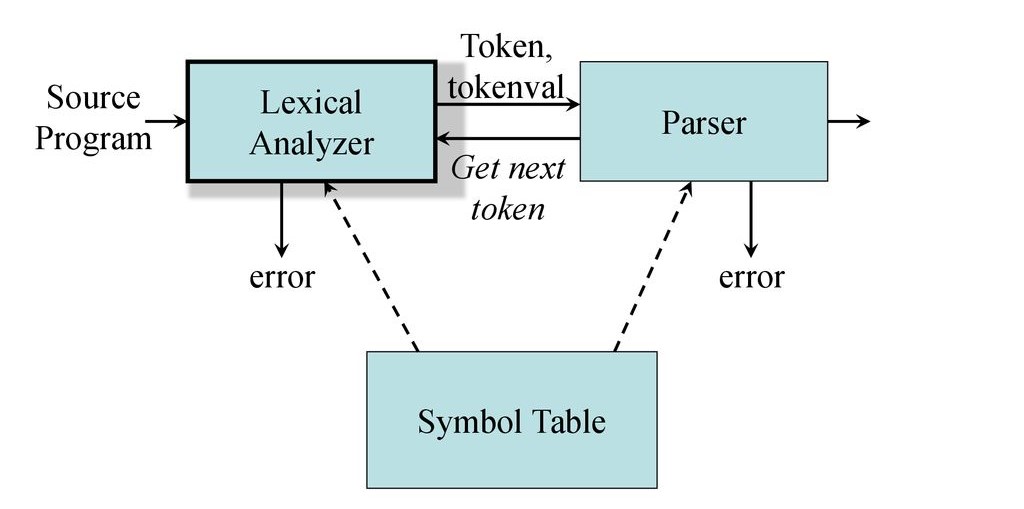
CSE471- Machine Learning CSE411- Algorithm Engineering CSE463- Introduction to Bioinformatics CSE309- Compiler Design CSE305- Computer Architecture CSE313- Operating Systems CSE405- Computer Security CSE321- Computer Networks CSE409- Computer Graphics CSE203- Data Structures and Algorithms MATH245- Statistics and Probability MATH247- Linear Algebra -
Higher Secondary School Certificate (HSC)
Notre Dame College, Dhaka
2018GPA: 5.00/5.00
- Board Talent Pool Scholarship
- Placed 8th in Dhaka Board
-
Secondary School Certificate (SSC)
St. Joseph Higher Secondary School
2016GPA: 5.00/5.00
- Board Talent Pool Scholarship
Technical Skills
-
Programming Languages
Python, C/C++, C#, Java, JavaScript, SQL, HTML/CSS, Bash, LATEX, MySQL, x86 Assembly, Bison/Flex
-
Libraries/Frameworks
PyTorch, Scikit-learn, Node.js, NS2, xv6, FastAPI, ReactJS, Wireshark
-
Tools/Platforms:
Docker, Git, Google Colab, kaggle, Visual Studio Code, Linux
Languages
English (Fluent), Bengali (Native)